Content:
◉ Introduction to Proteinase K
Proteinase K is a broad-spectrum, nonspecific, proteolytic enzyme widely used in molecular biology to inactivate protein contaminants in prepared samples. This endopeptidase remains active over a wide pH range (optimal between 6.5 and 9.5) and relatively high temperatures (optimal 50-65°C).
It is highly resistant protein, it will continue to function in the presence of chaotropic salts, detergents and in high temperatures offering an efficient and dependable method for most demanding applications.
Proteinase K, also known as protease K, is secreted by Tritirachium album Limber if this mold is grown in the presence of keratin, hence the designation "K", as the sole nitrogen source.
Proteinase K |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| CAS number | 39450-01-6 | ||||||||
| Molecular mass | 391.466 g/Mol | ||||||||
| Formula | C29H27N2O12P | ||||||||
| Synonyms | Endopeptidase K, EINECS 254-457-8 | ||||||||
Proteinase K = Protein (long chain of amino acids) + ase (suffix for enzyme) + K (keratin)
◉ Activity
Proteases are enzymes that catalyze the cleavage of a protein into amino acids, proteinase K is a broad-spectrum serine protease belonging to the subtilisin class (there are two types of serine proteases, chymotrypsin and subtilisin). This enzyme belongs to the family of S8 peptidases. The predominant site of cleavage is the peptide bond adjacent to the carboxyl group of aliphatic and aromatic amino acids with blocked alpha-amino groups.

Calcium ion is the cofactor of proteinase K enzyme, calcium ions do not affect the activity of the enzyme but contribute to its stability (thermostability and protects proteinase K from autolysis), if calcium ions are removed (e.g. by using EDTA), the stability of the enzyme is reduced, but the proteolytic activity is maintained.
Proteinase K digestion for nucleic acid purification is usually performed in the presence of EDTA (necessary to inhibit metal ion-dependent enzymes, such as nucleases). EDTA does not directly affect enzyme activity, it can impact the calcium and therefore reduce proteinase K activity to some extent.
Proteinase K and RNases are usually added together in lysis buffer because they form an efficient combination. RNase will break down contaminating RNA and Proteinase K will break down damaged proteins, DNases and RNases.
◉ Proteinase K stability
Proteinase K is stable over a wide range of pH (from 4 to 12) and temperature (from +37°C to +65°C). Increasing the reaction temperature from 37°C to 50-60°C can increase activity many-fold, by making its substrate cleavage sites more accessible, as can adding 0.5-1% of sodium dodecyl sulfate (SDS) or guanidinium chloride (3 M), guanidinium thiocyanate (1 M) and urea (4 M).
It has a shelf life of 12 months when stored in a dry place at 4–8°C, due to the fact that it is a very stable. Short term storage at ambient temperatures do not harm the enzyme’s activity and stability.
◉ Proteinase K uses
◉ Proteinase K is commonly used in molecular biology to digest proteins, remove contaminations, and inactivate DNases and RNases that would otherwise degrade a desired DNA or RNA sample. Proteinase k is suitable for this application because the enzyme is active in the presence of protein denaturing chemicals such as SDS and urea, chelating agents such as EDTA, hydrogen sulfide reagents and trypsin or chymotrypsin inhibitors.
◉ It is used for the destruction of proteins in cell lysates (tissues, cell culture cells) and for the release of nucleic acids
◉ It is also useful for characterizing proteins : Due to the cleavage specificity of proteinase K, characteristic fragments of proteins are obtained which are useful for determining the structure and function of proteins.
◉ This enzyme is very useful in the isolation of highly native and undamaged DNA or RNA, since most microbial or mammalian DNases and RNases are rapidly inactivated by the enzyme, especially in the presence of 0.5 at 1% SDS.
◉ How do you inactivate proteinase K ?
Proteinase K is inactivated by heating at 95°C for 10 minutes ( heating does not fully inactivate the enzyme, there will always be a small amount of activity remaining through this method.), trichloroacetic acid (TCA) or serine protease inhibitors AEBSF, PMSF or DFP (permanently inactivate proteinase K)
◉ What is the function of proteinase k in DNA extraction?
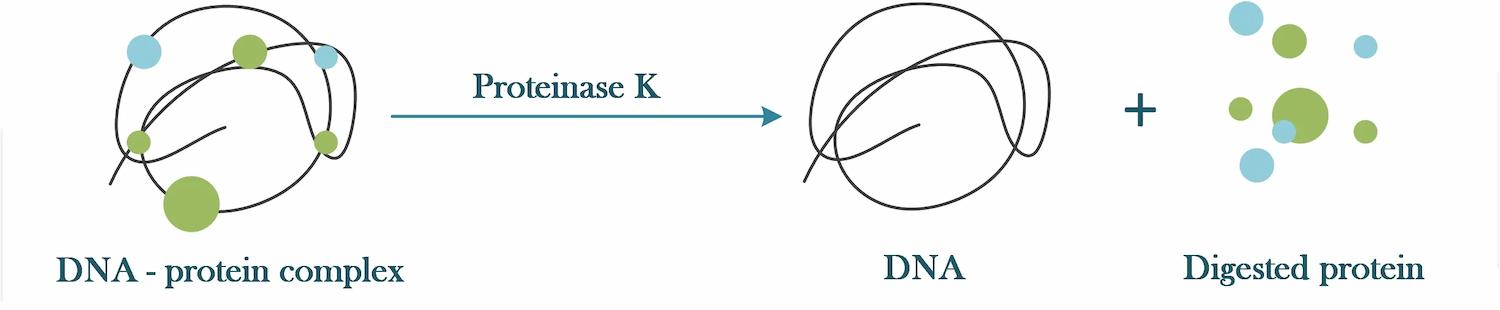
During the extraction of DNA (or nucleic acids in general), many contaminating proteins are present. Proteinase K is used in many DNA extraction protocols to digest these contaminating proteins.
Additionally, nucleases (enzymes that break down nucleic acids) may be present, the addition of proteinase K degrades these nucleases and protects the nucleic acids from nuclease attack.
◉ Frequently Asked Questions
Q : What is the protein concentration of Proteinase K, Molecular Biology Grade ?
R : The protein concentration of the enzyme is ~20 mg/ml and its unit activity is 800 U/ml.
Q : Why do we neutralize the proteinase K after incubation ?
R : It is inactivated after incubation to prevent potential digestion of the samples.
Q : WHAT ARE BUFFERS ACCORDING TO PROTEINASE K ACTIVITY?
R :
|
Buffer (pH 8.0, 50°C, 1.25 µg/ml protease K, 15 min incubation) |
Proteinase K activity (%) |
|
30 mM Tris·Cl |
100 |
|
30 mM Tris·Cl; 30 mM EDTA; 5% Tween 20; 0.5% Triton X-100; 800 mM GuHCl |
313 |
|
36 mM Tris·Cl; 36 mM EDTA; 5% Tween 20; 0.36% Triton X-100; 735 mM GuHCl |
301 |
|
10 mM Tris·Cl; 25 mM EDTA; 100 mM NaCl; 0.5% SDS |
128 |
|
10 mM Tris·Cl; 100 mM EDTA; 20 mM NaCl; 1% Sarkosyl |
74 |
|
10 mM Tris·Cl; 50 mM KCl; 1.5 mM MgCl2; 0.45% Tween 20; 0.5% Triton X-100 |
106 |
|
10 mM Tris·Cl; 100 mM EDTA; 0.5% SDS |
120 |
|
30 mM Tris·Cl; 10 mM EDTA; 1% SDS |
203 |