Summary :
What type of antibiotic is Polymyxin?
Polymyxins are antibiotics naturally produced by different species of Paenibacillus (Bacillus) polymyxa. Five chemical classes (A, B, C, D and E) are described, but only two compounds are used therapeutically :
- Polymyxin B
- Polymyxin E or Colistin
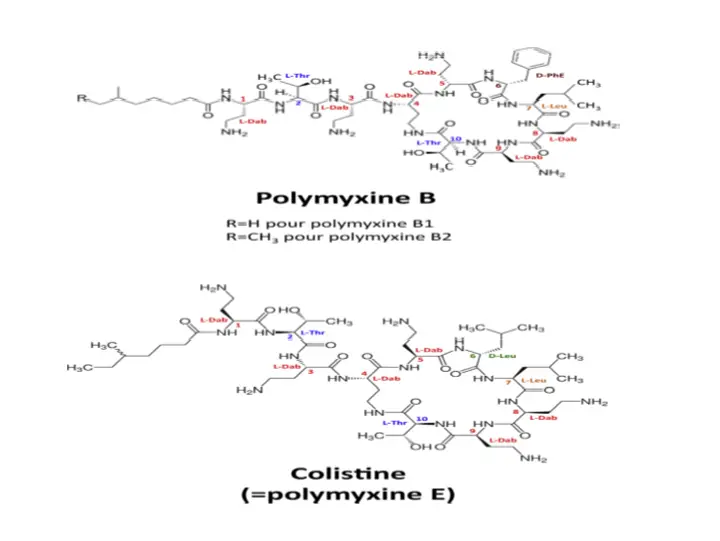
Structure of Polymyxin
Molecules belonging to the polymyxins family are large, with a molecular mass of approximately 1200 Da. They are cationic polypeptides consisting of a cycle of 7 amino acids and a tripeptide side chain to which a fatty acid is covalently linked.
This common chemical structure gives them both their hydrophilic property (thanks to the free amine groups of L-2,4-iaminobutyric acids) and their lipophilic property (thanks to their fatty acid and the amino acids in position 6 and 7 of the heptapeptide ring).
Polymyxin B differs from Polymyxin E by a single amino acid at position 6, where the D-phenylalanine of Polymyxin B is replaced by D-leucine in colistin . Their in vitro antibacterial activity is comparable but they are administered in two different forms by the parenteral route. While polymyxin B is administered directly in an active form, colistin is administered as a prodrug (colistimethate sodium) and therefore needs to be transformed in vivo into an active metabolite .
Pharmaceutical forms and dosage
There are two marketed pharmaceutical forms for colistin:
➣ colistin sulfate for oral use (COLIMYCINETM, 1,500,000 IU, tablet, Sanofi) and for topical use;
➣ colistimethate sodium (COLIMYCINETM, 1,000,000 IU, powder for solution for injection, Sanofi).
Colistin sulphate and colistimethate sodium are hardly absorbed from the gastrointestinal tract. This is why colistimethate sodium is used parenterally for the treatment of deep infections. Similarly, after intravenous (IV) injection, the distribution of colistin in the cerebrospinal fluid is relatively low, implying the use of this antibiotic by IV and intraventricular routes for the treatment of systemic infections. central nervous system.
Mechanism of action of polymyxin
The target of polymyxins is bacterial lipopolysaccharide (LPS), a component of the outer membrane of Gram-negative bacilli.
The mechanism(s) of action of polymyxins are not fully understood. However, according to data from the literature, polymyxins could act according to three distinct and concomitant modes leading to the death of the bacterium:
➀ Lysis of bacterial membranes (main route);
➁ Vesicle-vesicle contact;
➂ Formation of free radicals.
In addition, anti-toxin activity has been reported.
➀ Lysis of bacterial membranes
Lipopolysaccharide is composed of three domains :
➀ Lipid A located in the outer membrane of the bacterium.
➁ A central part (central core) oligosaccharide .
➂ A polysaccharide chain forming the O antigen.
Lipid A plays a major role in the mode of action of polymyxins. Indeed, the chains of fatty acids of lipid A allow the anchoring of polymyxins in the external membrane of the bacterium. Moreover, thanks to its negative charge, lipid A interacts with divalent cations (Ca2+ and Mg2+) present on the surface of the outer membrane. Its divalent cations then form bridges between the LPS molecules, thus allowing the overall stabilization of the outer membrane.
Since the affinity of LPS for polymyxins is greater than that of LPS for divalent cations (Ca2+ and Mg2+), an electrostatic interaction takes place initially between the LPS, negatively charged, and the antimicrobial polypeptide, positively charged . Polymyxins are therefore able to displace cations from their binding site destabilizing the structure of the outer membrane. After this initial step of electrostatic interaction, polymyxins insert into the outer membrane near lipid A via their N-terminal fatty acid chain.
The gaps thus formed allow the passage of hydrophobic molecules, small proteins and also facilitate the insertion of other polymyxin molecules. Eventually, this results in the formation of highly destabilized membrane zones through which a few polymyxin molecules can cross the outer membrane. Polymyxins are then able to lyse the cytoplasmic membrane of the bacterium, inducing bacterial lysis responsible for the bactericidal effect of its antibiotic molecules.
➁ Vesicle-vesicle contact
The outer membrane is composed of an inner phospholipid layer only and an outer layer containing mainly LPS, proteins and lipoproteins. After crossing the outer membrane, the polymyxins find themselves in the inter-membrane space where they are able to bind to the anionic phospholipids composing both the inner leaflet of the outer membrane and the outer leaflet of the inner membrane (or membrane cytoplasmic) of the bacterium. The exchange of lipids between the two membranes (external and internal) induces a loss of specificity in the composition of the membranes. This would lead to an osmotic imbalance responsible for the lysis of the bacteria
➂ Formation of free radicals
It has recently been demonstrated that polymyxins can induce the death of the bacterium via the accumulation of hydroxyl radicals (OH). Indeed, polymyxins are able to induce oxidative stress leading to the formation of reactive oxygen species (ROS) such as superoxide ions (O2), hydrogen peroxide (H2O2) and hydroxyl radicals (OH ) . The O2 ions would be produced during the crossing of the outer and inner membrane by the molecule of polymyxin, before being transformed into H2O2 by the superoxide dismutase (SOD) of the bacterium. H2O2 then oxidizes ferrous ions (Fe2+) to ferric ions (Fe3+) according to the Fenton reaction, inducing the formation of OH radicals. High levels of OH are then responsible for damage to DNA (breakage), proteins (oxidation) and lipids (oxidation), leading to the death of the bacteria.
Anti-toxin activitye
In addition to their direct bactericidal activity, polymyxins also have anti-endotoxin activity. Indeed, by binding to LPS, polymyxins also neutralize lipid A, a toxic compound allowing the anchoring of LPS in the outer membrane.